1. Introduction:
1.1. Introduction
Complement system in human body plays a imporatant role to provide first line defence against pathogen and any foreign bodies which have antigenic property (Noris & Remuzzi, 2013). It is a part of inner immunity and maintain a cross-talk with other immune cells and cytokines to provide a basic protection. Therefore body homeostasis is maintained by the activity of complement system. There are a lot of plasma and membrane proteins are involved in this sytem which work alternatively and make a barrier against foreign and altered cells (Merle et al., 2015). Cascade collaboration of plasma proteins which are involved in complement system are known to opsonize pathogens and activate inflammatory reactions. There are three different pathways of complement activation like classical pathway, alternative pathway and lectin pathway. These all pathway are bridged with a common terminal pathway (Merle et al., 2015). However though complement system is important enough to function against pathogen overexpression or insufficient expression of it may have detrimental effects on host. Complemet system has association with tumor progression and cancer too. Cancer which can be defined by an abnormal situation within the body where the tumors are not benign and can migrate from one place to another. In early twenty first century it was discovered that inflammation is associated with Cancer progression (Blackadar, 2016). Later studies suggested the activity of complement proteins in tumor micro-environment (Zhang et al., 2019). Several membrane proteins are there which after complement activation get activated and then control adaptive and humoral immunity, angiogenic processes, migration and mobilization of stem cells (Zhang et al., 2019).
Some complement regulators are also there which acts to regulate the expression of proteins of complement system. Complement regulators regulate either by inhibiting protease activity or by regulating convertase and Membrane Attacking Complex (Bayly-Jones, Bubeck & Dunstone, 2017). Sometimes regulator proteins bind with complement proteins as they have some specific domains. Therefore the expression of complement proteins are very well regulated in body by several means. There are two different types of complement regulators in body. One is soluble factors and another one is membrane bound. They modify and alter in complement proteins and ultimately maintain cellular homeostasis. CD 55 is such a complement regulatory proteins which is also known as decay-accelerating factors as it helps to inactivate complement system (Dho et al., 2018). CD 55 is a membrane bound protein. Several research have already been done to investigate the expression of CD 55 on lung, prostate, colorectal cancer, leukemia and cervical cancer (Dho et al., 2018). As CD 55 is anti-regulator of complement system it prevents to lyse cancer cells therefore it facilitates tumor progression and angiogenesis. Apart from that CD 55 also inhibits natural killer cells to perform therefore, it is a beneficial protein for cancer progression (Dho et al., 2018). Different cytokines like Tumor necrosis factor, interleukin 6 and interleukin 1 all are responsible for increased expression of CD 55. Different growth factors like EGF and mitogen are also responsible for excessive expression (Dho et al., 2018). Hematologic malignancy is a separate domain of cancer that includes cancer in white blood cells, bone marrow and lymph nodes. However there are four different types of hematologic malignancy like Acute Myloied leukemia, Acute lymphocytic Leucemia, Hodgkin, and non-hodgkin lymphoma. Though CD 55 is associated with all of these types of cancer, however there are differences regarding the genome splicing and expression of protein (Dho et al., 2018).
The main objective of the study is to make a comparison between the expression of CD 55 as a complement regulatory protein in four different haematological malignancies.
1.2. Background
Many studies have already been done to investigate the expression of different complement regulatory proteins in different types of cancer cells (Hara et al., 1992). This regulators are directly or indirectly associated with proliferation of progression of cancer cells by blocking apoptosis, and complement and natural killer cell mediated cell killing (Dho et al., 2018). Alteration in cytokine, cellular cross talk are associated with the activity of CD 55 which ultimately leads to malignancy. CD 55 is now been considered as a target of cancer. Several drugs and monoclonal antibody are used against CD 55 to reduce its’ activity (Dho et al., 2018).
1.3. Problem statement
Though CD 55 is majorly responsible with different types of cancer, it has significant role over haematological malignancies. Immune responsiveness is a very special clinical feature of blood malignancies (Dho et al., 2018).. Therefore different immunological functions, inflammation, action of antigen presenting cells are alternatively associated with haematological malignancies. As discussed above that CD 55 prevent complement protein to establish a barrier against foreign and neoplastic cells therefore CD 55 is a key target of haematological malignancies (Dho et al., 2018).. However the expression of CD 55 varies in different types of haematological cancer like Acute Myloied leukemia, Acute lymphocytic Leukemia, Hodgkin, and non-hodgkin lymphoma. There is no comparison study regarding that. Therefore it is leaving a room to make a review on comparison between expression of CD 55 in four different cancers.
1.4. Rationale
There are several individual studies have already been done on the role and function of CD 55 protein in different types of cancer progression like prostate cancer, lung cancer, hodking and non hodking lymphoma, Acute myloied leukemia, and Acute lymphocytic leukemia. In some of these cancer cells the CD 55 or the DAF factor remains activated due to the alteration in proten sequence due to polymorphism or because of the altered cell signalling pathway (Nakagawa et al., 2001). Therefore complement inactivation process remains activated for a longer time and the general immune action against the neoplastic cells becomes non-functional (Spendlove et al., 2006). The altered functionality of CD 55 protein depends on conformational change of protein. Polymorphism is responsible for alteration in DNA promoter activity therefore, if any upregulatory or downregulatory mutation is there the synthesis of RNA will be consecutive or diminished and abnormal protein production will be there in the cancer or altered self cell that will either prevent the activation of complement regulatory system and prevent natural killer cells to perform against the non-self cells or help to function complement cascade (Brodsky, 2014).
1.5. Aims and Objectives
The study aims to investigate the expression of CD 55 in four different haematological malignancies like Acute myeloid leukemia, Acute lymphocytic leukemia, Hodgkin lymphoma, and non Hodgkin lymphoma. Apart from that the relation between polymorphism, protein sequence alteration and expression variability in four different malignant cancers is also a conjoint objective of this study.
The major objectives of this study are,
- To investigate the sequence of CD 55 protein in four different haematological malignancies like AML, ALL, Hodgkin and Non Hodgkin lymphoma.
- To investigate the expression difference of CD 55 protein in above mentioned cancer.
- To investigate how the expression variability is linked to the cancer progression.
1.6. Structures of research
RQ.1. what is the sequence of CD 55 in normal cell?
RQ.1. How the sequence varies in AML, ALL, Hodgkin and non Hodgkin lymphoma due to polymorphism?
RQ.3. How the expression variability is linked to the sequence alteration in four different type of haematological malignancies?
2. Literature review
2.1. About haematological malignancies
Cancer is caused by genetic disorder. Mutation in proto oncogene turns it activated to oncogene and the mutation in tumor suppressor gene inactivates it permanently. Therefore normal cells achieve some abnormal changes that leads to the neoplastic alteration. Activation of oncogene gives the cells a potential to proliferate uncontrollably, and inactivation in tumor suppressor genes are responsible for the incapability of DNA repair protein to repair the changes due to polymorphism (Lee & Muller, 2010). Different types of cancers have been reported till death which are associated with different organs. However haematological malignancies are a particular type of cancer which involves immune cells like granulocytes, agranulocytes, bone marrow and cells of lymphatic system (Taylor, Xiao & Abdel-Wahab, 2017). Four major types of haematological malignancies are Acute myeloid leukemia, Acute lymphocytic leukemia, Hodgkin lymphoma, and non Hodgkin lymphoma (Taylor, Xiao & Abdel-Wahab, 2017)..
2.2. Different types of haematological malignancies:
Acute myeloid leukemia is most common form of leukocytes malignancy in adults. In United States almost twenty thousand people get affected with Acute myeloid leukemia in every year (De Kouchkovsky & Abdul-Hay, 2016). Chromosomal translocation or mutation in genome of Hematopoietic stem cells results in inappropriate cell differentiation therefore the accumulation of poorly differentiated myeloid cells takes place. Some clinical symptoms are also there which are specific for acute myeloid leukemia like thrombocytopenia and anemia, due to lack of proper precursor cells (De Kouchkovsky & Abdul-Hay, 2016). The initiation of acute myeloid leukemia had been reported in soft tissues in bone marrow (De Kouchkovsky & Abdul-Hay, 2016). Mainly the white blood cells precursors get affected by acute myeloid leukemia due to the genetic changes and alteration in some oncogenic proteins (De Kouchkovsky & Abdul-Hay, 2016). However from bone marrow the neoplastic immature white blood cells move from bone marrow to circulatory system which includes both blood circulatory system and lymphatic system. The precursor of red blood cells and platelets also get affected to this. Chromosomal translocation and random mutation are associated with cancerous heterogeneity of immatured blast cells. Cytogenetics study had been done to detect the mutation hotspot in AML. Genes like KIT, NPM 1, RAS, ERG, DNMT, CBL, IDH, and TET 2 had been found to have relation with Acute myeloid leukemia (Alrajeh, Abalkhail & Khalil, 2017). Mutations in different chromosomal positions had been detected by karyotyping. Some of the mentioned genes have oncogenic activity where some have biochemical alterations activity. Acute lymphocytic leukemia is majorly associated to lymphoid progenitor cells (Terwilliger & Abdul-Hay, 2017). From lymphoid progenitor cells mature lymphocytes like B and T lymphocytes produce and these cells act as a defensive barrier against foreign pathogen and non-self cell. However when the lymphoid progentitor cells are mutated then the differentiation of it to precursor cells are abnormal and as a result a production of a huge amount of malignant lymphocytes takes place. The process intitatially begins at stromal region of bone marrow as this is the place where ancestor cells give rise to B and T lymphocytes (Terwilliger & Abdul-Hay, 2017). With the altered functionality then they move to circulatory pathway and then achieve the potential to invade other tissues. Like Acute myeloid leukemia, Acute lymphocytic leukemia is also caused by random genetic mutation and chromosomal translocation. Mutation in some specific genes cause acute lymphocytic leukemia. Chromosomal translocation which had been reported till date include t(12;21) [ETV6-RUNX1], t(9;22) [BCR-ABL1], t(1;19) [TCF3-PBX1] (Safaei et al., 2013). Apart from that very recently a different type of translocation that is very common in case of chronic myeloid lymphoma had also been found in case of acute lymphocytic leukemia too. This rearrangement is more or less similar to that of Philadelphia chromosomal translocation that helps to make ALL-BCR complex (Safaei et al., 2013). However ALL-BCR complex formation does not occur in acute lymphocytic leukemia. It has been shown that more or less 80% of the cases of Ph like Acute lymphocytic leukemia the factors are responsible with deletions in a major transcription factors that are associated with the development of B cell. It includes Zinc finger motif of IKAROS family, E2A oncogenic transcription factor, Early B cell factor, and PAX 5. Several mutations which are associated with kinase activation are also common in 90% of Ph like Acute lymphocytic leukemia (Safaei et al., 2013). Rearrangement of ABL 1, JAK 2, EPOR, CRLF 2 are the examples of it. All of these rearrangements are associated with cancer progression and development of immatured B lymphocytes (Safaei et al., 2013).. There immune property also gets altered with the genotypic alteration and they are no longer to elucidate immunity against non-self cells. Hodgkin lymphoma is a specific type of haematological malignancy which originates from precurson B lymphocytes (Shanbhag & Ambinder, 2018). It can be characterized by the defective expression progression of B cell. Epigenetic transition and downregulation in transcription factors are associated with the clonal proliferation and development of Reed Sternberg and Hodgkin cells (Shanbhag & Ambinder, 2018). The progression of cancer is associated with the chromosomal alteration and genetic mutation. Mutation study had been confirmed that the phenotypic alteration of lymphocytes are associated with genomic heterogeneity (Shanbhag & Ambinder, 2018). Signaling molecules which are involved in cellular cross-talk have been found to show altered activities in Hodgkin lymphomas. GM-CSF/IL-3, JAK/STAT, CBP/EP 300, NF-kappaB are known to alter the pathway of immunoglobulin chain rearrangement and activities of RAG1/RAG2 recombinase (Shanbhag & Ambinder, 2018). Mutational hotspot have also been detected in gene like BTK, CARD 11, and BCL 10 (Shanbhag & Ambinder, 2018). These all are associated to the development of BCR and cancer progression (Shanbhag & Ambinder, 2018). Non Hodgkin lymphoma is an another type of haematological malignancies involve white blood cells, more specifically the agranunolocytes or lymphocytes. It can be classified into both B cell and T cell lymphomas.
2.3. Association of CD 55 with progression of cancer
CD 55 is a complementation regulatory protein which was isolated primarily from human erythrocytes. It is a GPI linked membrane protein which binds with a specific protein known as CD 97 (Dho et al., 2018). Though the major role of it to inhibit the activity of complentation system it has other association in cancer progression too (Dho et al., 2018).. This membrane protein is prevalent in blood cells, stromatal cells, epithelial and endothelial cells. Soluble CD 55 is also been found in urine, plasma, tears, saliva, and different types of body fluids. The CD 55 protein is encoded from a gene which is located on the short arm on human chromosome number 1 (Dho et al., 2018).. The locus is also can be identified as chromosome 1q32. 11 exons are there in the 40 kilo base pair long fragments of the gene from where the matrix protein is encoded after alternative splicing (Dho et al., 2018). Seven different isotypes of the protein have been detected till the date in normal cells. The mature CD 55 protein is a 43 kDa protein (Dho et al., 2018). However it also varies in different cell types. The range varies in 50 kDa to 100 kDa. Different alternative splicing is responsible for the variable molecular weight and size of the protein. CD 55 is mainly a complement inactivation protein. It act by inhibiting the early activation of complement pathway. It accelerates and promotes the degradation of C3 convertase which is an activator of complement system and has a regulatory role to maintain the complement activation cascade (Dho et al., 2018). Apart from that it also inhibits the molecular assembly of the protein C3 convertase by dissociating the complex of fixed C3b from Bb protein and thus it ultimately blocks the activation of complement pathway. CD 55 has also an interesting role that it can downregulates the activity of T lymphocytes and this is generally done via an pathway that is not linked with complement pathway. A receptor specific for CD 55 protein is there in membrane of different cells and this receptor is known as CD 97 (Dho et al., 2018). CD 97 is prevalent in T and B lymphocytes and the interaction is associated with the development of lymphocytes. However when the morphology of protein gets altered the receptor binding gets changed and results in abnormal functionality (Dho et al., 2018).. It has been reported that when there is overexpression of such negative regulator of complement system in cells, it results in complement dependent cytotoxicity in cancer cells. Therefore the restriction of therapeutic potential of antibodies and drugs which are used to treat cancer are assumed to be fixed by blocking the the protein like CD 55, CD 97, and others complement regulatory protein which are associated with cancer progression.
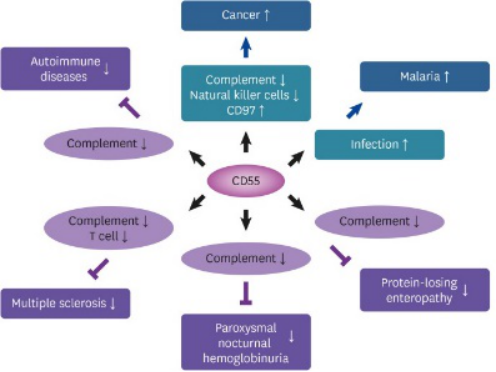
Figure 1: Activity of CD 55 protein
Source: (Dho et al., 2018).
In cancer cell the factors have already been studied which are associated with upregulation of CD 55 protein. Several molecular weight factors like different growth factors, interleukenes and cytokines upregulates the expression of complement regulatory protein. IL-1 β and IL-4 are two interleukenes which have been found to increase the concentration of CD 55 protein in epithelial cells of human intestine (Dho et al., 2018).. Apart from that Tumor necrosis factor, Interleukene 6 and Interleukene 4 are known to increase the expression of CD 55 in hepatoma cells of human (Dho et al., 2018).. Mitogen activated protein kinase which is also written as MAPK pathway had been found to have an association with CD 55 overexpression (Dho et al., 2018).. Epithelium growth factor acts as an triggering ligand in that case. Endothelial growth factor is another growth factor which by interacting with tyrosine kinase receptor increase the expression of the complement regulatory factors (Dho et al., 2018).. However the expression of CD 55 varies in haematological malignancies. CD 55 is responsible for the growth and development of hematopoietic steam cells from which the blood cells are produced. The expression of CD 55 in red blood and white blood cells of acute myeloid leukemia is lessly detected. However the pathway of the downregulation is not clear yet.
3. Methodology:
3.1. Sequence identification of CD 55 human protein
Amino acid of protein and nucleotide sequence of amino acid is generally performed in laboratory. However the sequenced data of different proteins and genomes are also available at proteiomics and genomics database site. Database sites like NCBI, UNIPORT, EXPACY, HSLS are there and the sequence of a protein motif, domain and even of a complete protein can be obtained from the above mentioned sites. Genomic data are also available at NCBI. For this experiment the secondary structure of the protein CD 55 and the structure of matured protein was obtained from UNIPORT.ORG.
3.2. Genomic sequence identification of CD 55 human protein
NCBI was used to obtain the genomic sequence of CD 55 which is located on the short arm of chromosome 1. The sequence and the pattern of exon and intron was available in both FASTA and genbank format. However as the FASTA sequence is long enough the graphical data of gene sequence was obtained from the site.
3.3. Findings of CD 53 protein and genomic sequence in haematological malignancies
To find the whether there is any change in the sequence of CD 55 protein in acute myeloid leukemia, Acute lymphocytic leukemia, Hodgkin lymphoma, and non Hodgkin lymphoma NCBI was used. However adequate data regarding the genomic and proteomic sequence of CD 55 in the above mentioned cancer cell lines were not available there in the site. Also some other sites like UNIPORT and EXPACY were searched but the result was same.
3.4. Finding the pattern of polymorphism in CD 55 genome
To find the pattern of polymorphism in the genome of CD 55 in four different haematological malignancies several articles were searched. Some papers were able to give idea regarding the pattern of polymorphism and the site of mutation in the genome of CD 55. Therefore the articles are used to correlate the information with the genomic sequence of CD 55 in normal cell.
3.5. Finding the expression of CD 55 membrane protein in four different cancer cell line.
Secondary data were used to know the expression of CD 55 in different haematological malignancies. Some experiments were done to detect the expression of CD 55 in ALL, CML and AML. Therefore the articles are also used to obtain the data regarding the expression of CD 55.
4. RESULTS
4.1. Findings of protein sequence
UNIPORT.ORG was accessed to get the protein sequence and structure of human CD 55 in normal cells. Reactome had shown four different databases like R-HAS-373080, R-HAS-6798695, R-HAS-6807878, and R-HAS-977606. All of these have different functions. The last one have complement cascade regulatory activities. Where the others are associated with cellular functions like neutrophil degranulation and anterograde transport.
However the pattern of secondary structure of CD 55 and the structure of the structure of matured protein were obtained from the site.
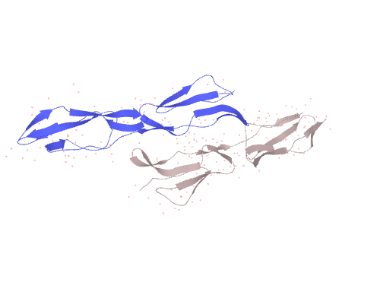
Figure 2: The X-ray crystallography structure of human CD 55 (Isotype 1)
Source: Uniport.org
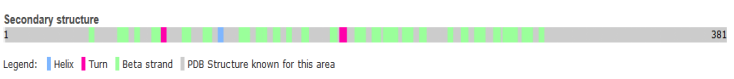
Figure 3: Secondary structure of human CD 55
Source: Uniport.org
4.2. Cannonical sequence of CD 55 (Isoform 2)
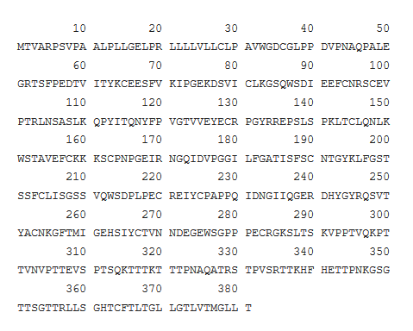
Figure 4: Cannonical sequence of CD 55 protein
Source: Uniport
4.3. Findings of polymorphism in CD 55 sequence in haematological malignancies
Adequate data regarding the sequence of CD 55 was not available in database. Therefore the pattern of polymorphism was studied from articles. Though information regarding polymorphism in CD 55 genome of lymphoma was not found, a mutation study was done to investigate the effect of polymorphism in CD 55 genome and how it is associated with lung cancer susceptibility (Zhang et al., 2017). A variant type of CD 55 rs 2564978 was found in lung cancer and it was shown that it had the potential to increase the susceptibility as the mutation increased the activity of the promoter of the gene (Zhang et al., 2017). The study was not based on haematological lymphoma, but as the functionality of the protein is more or less similar in every cell therefore it is predicted that mutation in different places would upregulate or downregulates the activity of CD 55.
4.4. Expression of CD 55 in haematological malignancies
An experiment was performed to detect the expression of different complement regulatory proteins on different haematological malignancies (Guc et al., 2000). That study includes four different cancer cell lines of Chronic myeloid leukemia, Acute lymphocytic leukemia, Non Hodgkin lymphoma, and acute myeloid leukemia. In that experiment the expression of DAF was measured with Fluorescence activated cell sorting method by using anti-CD 55 antibody. Expression of some others complement proteins like CR1 and MCP was checked in that experiment but as the main objective of this research is associated with CD 55, the expression of DAF would only be focused. Result suggested that one of the total Acute myeloid leukemia cell line and Three of the total Non hogkin lymphoma cell line had shown negative expression for CD 55. Therefore it was concluded that some of the haematological malignancies like ALL and NHL lacks CD 55, therefore they are no longer capable to inhibit complementation. However they have other mechanisms to escape immune system. The amount of CD 55 was more or less 40% deficient in every cell lines of haematological malignancies than that of the normal cell lines. Therefore it can be said that the expression of CD 55 in haematological malignancies are lesser than the normal cells. When some haematological malignancies show low expression level for CD 55, others are lack of it.
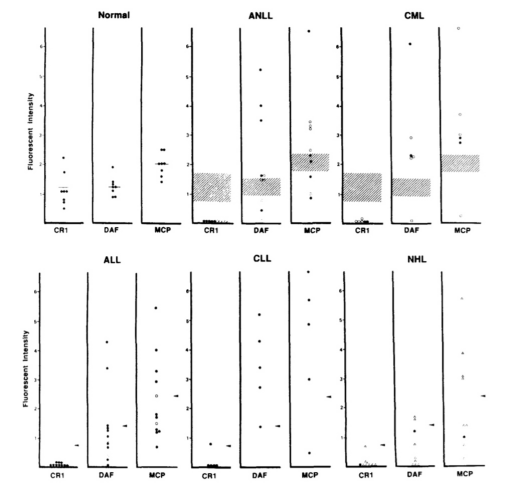
Figure 5: Expression of complement regulatory protein in haematological malignancies
Source: (Guc et al., 2000)
5. Discussion:
CD 55 is a complement regulatory protein and the expression of it alters in different cells. Not every cells in human body contains same amount of CD 55. Therefore it can be said that the expression of CD 55 is associated with different transcription activators and different ligands. Though the major purpose of it is to regulate the complement activation cascade, it is also responsible to control the progression of cancer. Many researches had already been done to investigate the relation between CD 55 expression and lung cancer, prostate cancer, and esophageal cancer. As the main objective of this paper was to investigate the significance and difference in expression of CD 55 in four different cancer database and article based study have been followed to find the answers. Though the sequence of human CD 55 protein was possible to obtained there was data insufficiency regarding the expression of CD 55 in four different haematological malignancies. The polymorphism in DNA sequence of CD 55 mentions that there is a relation between the mutation and altered protein expression. In maximum cases Acute myeloid leukemia and Non Hodgkin lymphoma cancer cells do not contain membrane CD 55 proteins though the Acute lymphocytic leukemia cancer cells do contain CD 55 in lower level. Therefore it is clear from this portion that there is a relation between the progression of cancer and the expression of CD 55 in haematological lymphoma which is need to be explored and the factors which are associated with the downregulation of CD 55 protein in haematological lymphomas are also need to be studied to proceed the future study.
References:
Alrajeh, A. I., Abalkhail, H., & Khalil, S. H. (2017). Cytogenetics and molecular markers of acute myeloid leukemia from a tertiary care center in Saudi Arabia. Journal of Applied Hematology, 8(2), 68.
Bayly-Jones, C., Bubeck, D., & Dunstone, M. A. (2017). The mystery behind membrane insertion: a review of the complement membrane attack complex. Philosophical Transactions of the Royal Society B: Biological Sciences, 372(1726), 20160221. https://doi.org/10.1098/rstb.2016.0221
Beatty, G. L., & Gladney, W. L. (2015). Immune escape mechanisms as a guide for cancer immunotherapy. Clinical cancer research : an official journal of the American Association for Cancer Research, 21(4), 687–692. https://doi.org/10.1158/1078-0432.CCR-14-1860
Blackadar C. B. (2016). Historical review of the causes of cancer. World journal of clinical oncology, 7(1), 54–86. https://doi.org/10.5306/wjco.v7.i1.54
Brodsky R. A. (2014). Paroxysmal nocturnal hemoglobinuria. Blood, 124(18), 2804–2811. https://doi.org/10.1182/blood-2014-02-522128
De Kouchkovsky, I., & Abdul-Hay, M. (2016). Acute myeloid leukemia: a comprehensive review and 2016 update. Blood cancer journal, 6(7), e441-e441.DOI: 10.1038/bcj.2016.50
Dho, S. H., Lim, J. C., & Kim, L. K. (2018). Beyond the Role of CD55 as a Complement Component. Immune network, 18(1), e11. https://doi.org/10.4110/in.2018.18.e11
Guc, D., Canpınar, H., Kucukaksu, C., & Kansu, E. (2000). Expression of complement regulatory proteins CR1, DAF, MCP and CD59in haematological malignancies. European journal of haematology, 64(1), 3-9. https://doi.org/10.1034/j.1600-0609.2000.80097.x
Hara, T., Kojima, A., Fukuda, H., Masaoka, T., Fukumori, Y., Matsumoto, M., & Seya, T. (1992). Levels of complement regulatory proteins, CD35 (CR1), CD46 (MCP) and CD55 (DAF) in human haematological malignancies. British journal of haematology, 82(2), 368–373. https://doi.org/10.1111/j.1365-2141.1992.tb06431.x
Lee, E. Y., & Muller, W. J. (2010). Oncogenes and tumor suppressor genes. Cold Spring Harbor perspectives in biology, 2(10), a003236. https://doi.org/10.1101/cshperspect.a003236
Merle, N. S., Church, S. E., Fremeaux-Bacchi, V., & Roumenina, L. T. (2015). Complement system part I–molecular mechanisms of activation and regulation. Frontiers in immunology, 6, 262. https://doi.org/10.3389/fimmu.2015.00262
Nakagawa, M., Mizuno, M., Kawada, M., Uesu, T., Nasu, J., Takeuchi, K., … & Tsuji, T. (2001). Polymorphic expression of decay‐accelerating factor in human colorectal cancer. Journal of gastroenterology and hepatology, 16(2), 184-189. DOI: 10.1046/j.1440-1746.2001.02418.x
Noris, M., & Remuzzi, G. (2013). Overview of complement activation and regulation. Seminars in nephrology, 33(6), 479–492. https://doi.org/10.1016/j.semnephrol.2013.08.001
Roma-Rodrigues, C., Mendes, R., Baptista, P. V., & Fernandes, A. R. (2019). Targeting Tumor Microenvironment for Cancer Therapy. International journal of molecular sciences, 20(4), 840. https://doi.org/10.3390/ijms20040840
Safaei, A., Shahryari, J., Farzaneh, M. R., Tabibi, N., & Hosseini, M. (2013). Cytogenetic findings of patients with acute lymphoblastic leukemia in fars province. Iranian journal of medical sciences, 38(4), 301–307.
Sarma, J. V., & Ward, P. A. (2011). The complement system. Cell and tissue research, 343(1), 227–235. https://doi.org/10.1007/s00441-010-1034-0
Shanbhag, S., & Ambinder, R. F. (2018). Hodgkin lymphoma: a review and update on recent progress. CA: a cancer journal for clinicians, 68(2), 116-132. https://doi.org/10.3322/caac.21438
Spendlove, I., Ramage, J. M., Bradley, R., Harris, C., & Durrant, L. G. (2006). Complement decay accelerating factor (DAF)/CD55 in cancer. Cancer immunology, immunotherapy, 55(8), 987. DOI: 10.1007/s00262-006-0136-8
Taylor, J., Xiao, W., & Abdel-Wahab, O. (2017). Diagnosis and classification of hematologic malignancies on the basis of genetics. Blood, 130(4), 410–423. https://doi.org/10.1182/blood-2017-02-734541
Terwilliger, T., & Abdul-Hay, M. (2017). Acute lymphoblastic leukemia: a comprehensive review and 2017 update. Blood cancer journal, 7(6), e577. https://doi.org/10.1038/bcj.2017.53
Zhang, R., Liu, Q., Li, T., Liao, Q., & Zhao, Y. (2019). Role of the complement system in the tumor microenvironment. Cancer Cell International, 19(1), 300. https://doi.org/10.1186/s12935-019-1027-3
Zhang, Y., Zhang, Z., Cao, L., Lin, J., Yang, Z., & Zhang, X. (2017). A common CD55 rs2564978 variant is associated with the susceptibility of non-small cell lung cancer. Oncotarget, 8(4), 6216. doi: 10.18632/oncotarget.14053